DNA extraction is a fundamental process in molecular biology, enabling scientists to isolate genetic material from various biological samples. Among the numerous methods available, silica bead-based DNA extraction has gained widespread adoption due to its efficiency and reliability. This technique relies on the principle of DNA binding to silica surfaces under specific chemical conditions, with pH playing a critical role in determining the success of the binding step. Understanding the pH requirements is essential for optimizing DNA yield and purity, which directly impacts downstream applications such as PCR, sequencing, and forensic analysis. In this article, we will explore the scientific basis of pH-dependent DNA binding, its practical implications across different sample types, and how it compares to other extraction technologies.
Technical Principles of Silica Bead DNA Binding
The binding step in silica bead DNA extraction is governed by the interaction between negatively charged DNA molecules and positively charged silica surfaces. This process is highly dependent on pH, as it influences the ionization states of both DNA and silica. At a pH value typically between 5.0 and 6.5, the silica surface becomes positively charged due to protonation, while DNA remains negatively charged because of its phosphate backbone. This charge difference facilitates electrostatic attraction, allowing DNA to adsorb onto the silica beads. If the pH is too high, the silica surface may lose its positive charge, reducing binding efficiency. Conversely, if the pH is too low, DNA may denature or become insoluble, leading to poor recovery.
Beyond electrostatic forces, the binding buffer often contains chaotropic salts, such as guanidine hydrochloride, which disrupt hydrogen bonding and enhance DNA-silica interactions. The optimal pH range ensures that these salts function effectively without compromising DNA integrity. For instance, a pH of around 6.0 is commonly used in commercial kits to maximize binding capacity while minimizing co-precipitation of impurities like proteins or polysaccharides. This precise control is crucial for obtaining high-quality DNA that is free from inhibitors, making it suitable for sensitive applications like next-generation sequencing. Researchers can refer to specialized protocols for challenging samples, such as those involving forensic bone extracts, where pH adjustments may be necessary to address inhibitors.
Comparison of DNA Extraction Technologies
Silica bead-based extraction is one of several methods used in modern laboratories, each with distinct advantages and limitations. Column-based systems, for example, employ a similar silica membrane but are often limited by lower binding capacity and slower processing times. In contrast, magnetic bead methods use functionalized particles that can be manipulated with magnets, offering higher throughput and automation potential, though at a greater cost. Solution-based techniques, such as phenol-chloroform extraction, provide high purity but involve toxic chemicals and longer hands-on time. When evaluating these methods, factors like Cracking efficiency, DNA purity, scalability, and cost must be considered. For high-volume applications, such as clinical genetic testing, magnetic bead kits may be preferred due to their compatibility with automated platforms.
According to a study published in Nature Methods, magnetic bead-based extraction now accounts for approximately 40% of sample preparation in next-generation sequencing workflows, largely due to its scalability and consistency. However, silica bead methods remain popular for their balance of cost-effectiveness and performance, especially in resource-limited settings. The choice of technology often depends on the specific requirements of the downstream application. For instance, PCR-based assays may tolerate lower purity DNA, while NGS demands high-molecular-weight DNA with minimal fragmentation. Understanding these nuances helps laboratories select the most appropriate kit, whether for microbial research or food safety testing.
Industry Insights and Emerging Trends
The DNA extraction market is evolving rapidly, with emerging technologies like "extraction-free" direct PCR challenging traditional kit-based methods. These approaches bypass purification steps altogether, using specialized buffers to lyse cells and directly amplify DNA. While this reduces time and cost, it is not suitable for all sample types, particularly those with high inhibitor content, such as soil or complex tissues. The rise of direct PCR has prompted kit manufacturers to optimize their binding buffers for faster protocols without sacrificing purity. For example, some companies now offer rapid silica bead kits that integrate pre-lysed samples into a streamlined workflow, ideal for forensic buccal swabs or point-of-care diagnostics.
Another trend is the adoption of ISO standards, such as ISO 18385 for forensic DNA extraction, which emphasizes contamination control and reproducibility. These guidelines often specify pH ranges for binding buffers to ensure consistency across laboratories. As the demand for portable and field-deployable extraction methods grows, pH-stable formulations are being developed to withstand variable environmental conditions. This is particularly relevant for environmental water sampling, where pH fluctuations can occur naturally. By aligning with these standards, manufacturers demonstrate a commitment to quality, which is critical for applications in regulated industries like healthcare and forensics.
Sample Type Adaptation and pH Optimization
Different sample types present unique challenges for DNA extraction, necessitating adjustments in pH and buffer composition. Blood samples, for instance, contain hemoglobin and other PCR inhibitors that require a binding pH at the lower end of the optimal range (around 5.5) to enhance selectivity. In contrast, plant tissues often have high levels of polysaccharides and polyphenols, which can co-bind with DNA if the pH is not carefully controlled. For such samples, a slightly higher pH (e.g., 6.5) may improve purity by reducing non-specific interactions. Protocols for plant tissue extraction frequently include additional steps, such as CTAB pretreatment, to remove these contaminants before binding.
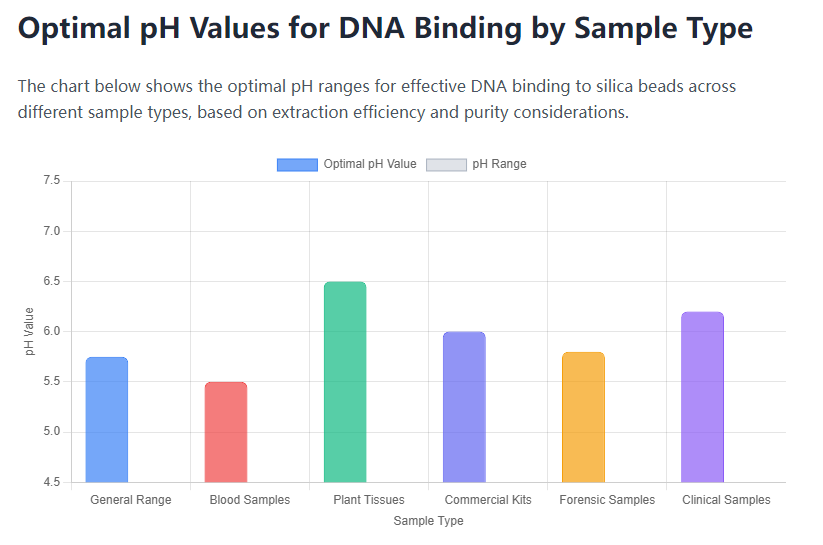
Forensic samples, like aged bloodstains or hair shafts, often contain degraded DNA and inhibitors from environmental exposure. Here, the binding pH must be optimized to recover low-abundance DNA while minimizing background interference. Similarly, microbial cultures from soil or water may require pH adjustments to account for extracellular polymers or humic acids. In clinical settings, samples like saliva or swabs benefit from standardized pH conditions to ensure reproducibility across batches. For example, kits designed for infectious disease testing often include buffering agents to maintain consistent pH during binding, regardless of sample variability. By tailoring the pH to the sample matrix, users can achieve higher yields and purities, facilitating reliable downstream analysis.
Downstream Applications and DNA Quality Requirements
The quality of DNA extracted via silica bead methods directly influences its suitability for various applications. PCR and qPCR, for instance, require DNA free from inhibitors like salts or organic compounds, which can be achieved by optimizing the binding pH to reduce co-purification. Next-generation sequencing, on the other hand, demands high-molecular-weight DNA with minimal fragmentation, necessitating a binding step that preserves integrity. A pH that is too acidic can cause DNA shearing, while an alkaline pH may lead to depurination. Therefore, most commercial kits target a neutral to slightly acidic pH (6.0–7.0) for binding, balancing stability and efficiency.
For applications such as molecular cloning or enzyme digestion, DNA purity is paramount, as contaminants can interfere with enzymatic reactions. Silica bead kits with optimized pH conditions often include wash steps to remove residual impurities, ensuring compatibility with these sensitive techniques. In forensic STR analysis, DNA must be of sufficient quality and quantity to generate reproducible profiles, making pH-controlled binding critical for success. Users working with challenging samples, such as semen stains, should verify that the binding pH aligns with their downstream workflow. By matching the extraction method to the application, laboratories can avoid costly repeats and ensure accurate results.
Purchasing Guide for DNA Extraction Kits
When selecting a DNA extraction kit, several key parameters should be evaluated, including yield, purity, speed, cost, and certification. Yield refers to the amount of DNA recovered, which can vary based on the binding capacity of the silica beads. Purity is typically assessed using absorbance ratios (e.g., A260/A280), with values near 1.8 indicating minimal protein contamination. Speed is a consideration for high-throughput labs, where rapid binding and elution steps can streamline workflows. Cost encompasses not only the kit price but also the expenses associated with instrumentation and labor. Certifications, such as ISO 13485 for medical devices, provide assurance of quality and reliability.
For instance, a kit designed for silica bead DNA extraction may offer a balance of these factors, making it suitable for diverse applications. However, users should confirm that the binding pH is specified and validated for their sample type. In research settings, flexibility may be prioritized, while clinical or forensic labs often require kits that comply with regulatory standards. Additionally, compatibility with automated systems can influence the choice, as seen in kits for blood research that integrate with robotic platforms. By carefully assessing these criteria, buyers can identify kits that meet their specific needs without compromising on performance.
Expert Recommendations and Best Practices
To achieve consistent results with silica bead DNA extraction, experts recommend calibrating the binding pH using standard buffers and verifying it with pH strips or a meter. For samples with inherent variability, such as soil or forensic evidence, performing a pilot extraction can help determine the optimal conditions. It is also advisable to use fresh binding buffers, as pH can drift over time due to environmental factors. When extracting DNA from plants, incorporating a polyvinylpyrrolidone (PVP) step can further reduce polyphenol interference, complementing pH optimization.
In cases where inhibition persists, increasing the wash stringency or incorporating a pre-binding cleanup may be necessary. For example, protocols for soil DNA extraction often include a centrifugation step to remove particulate matter before adjusting the pH for binding. Additionally, users should follow manufacturer guidelines for buffer storage and handling to maintain pH stability. By adhering to these best practices, laboratories can enhance the reliability of their DNA extraction processes, supporting applications ranging from basic research to diagnostic testing.
Conclusion and Decision-Making Tool
Understanding the pH requirements for the binding step in silica bead DNA extraction is essential for obtaining high-quality DNA. This parameter influences everything from yield and purity to compatibility with downstream applications. By considering factors such as sample type, downstream use, and regulatory requirements, users can select kits that align with their goals. For instance, a forensic lab might prioritize a kit with demonstrated efficacy on hair samples, while a clinical lab may seek one validated for oncology testing.
To assist in this process, ask the following questions when choosing a DNA extraction kit: What is the primary sample type? What downstream application will the DNA be used for? What is the available budget and throughput requirement? Are there any certification or regulatory standards to meet? How does the binding pH compare to optimal ranges for your samples? By addressing these points, you can make an informed decision that ensures successful DNA isolation for your specific needs.
