Silica bead DNA extraction technology represents a cornerstone methodology in modern molecular biology, particularly within high-throughput laboratory environments where efficiency, consistency, and scalability are paramount. These extraction systems leverage the fundamental binding properties of silica surfaces to selectively capture nucleic acids from complex biological mixtures while efficiently removing contaminants that could inhibit downstream applications. The inherent scalability of silica bead-based protocols has positioned this technology as a preferred solution for laboratories processing hundreds to thousands of samples daily, including genomic research facilities, clinical diagnostic laboratories, and forensic science centers. Understanding the performance characteristics, optimization strategies, and implementation considerations for silica bead systems enables laboratories to maximize their investment while achieving reliable, reproducible results across diverse sample types and applications.
Fundamental Principles of Silica-Based Nucleic Acid Binding
The molecular interaction between DNA and silica surfaces forms the foundation of this extraction methodology, relying on chaotropic salts that disrupt water structure and promote nucleic acid adsorption to the silica matrix. Under high salt conditions, the hydration shell surrounding DNA molecules is disrupted, enabling the phosphate backbone to interact directly with silicon dioxide particles through hydrogen bonding and van der Waals forces. This binding occurs preferentially over proteins and other cellular contaminants, providing the basis for selective nucleic acid purification. The binding efficiency varies with pH, salt concentration, and silica surface properties, with optimal conditions typically occurring at pH ≤7.5 and chaotropic salt concentrations exceeding 4M.
Silica bead technology improves upon traditional column-based silica methods by increasing the available surface area for DNA binding and facilitating more efficient mixing with sample lysates. The spherical geometry and uniform size distribution of high-quality silica beads maximize contact with nucleic acids in solution while minimizing bead aggregation that could reduce binding capacity. After DNA capture, washing steps employ ethanol-containing buffers that maintain the chaotropic conditions necessary to keep DNA bound while removing contaminants. The final elution step uses low-ionic-strength buffers or water that rehydrates the DNA molecules, disrupting their interaction with the silica surface and releasing pure nucleic acids into solution.
Throughput Advantages in Automated Laboratory Environments
High-throughput laboratories benefit significantly from the compatibility of silica bead systems with liquid handling automation, enabling parallel processing of 96 or 384 samples in standard microplate formats. The bead-based methodology eliminates the centrifugation steps required for column-based systems, instead relying on magnetic separation or filtration to isolate beads between processing steps. This transition from sequential to parallel processing reduces hands-on time from several hours to minutes while improving inter-sample consistency through standardized robotic liquid handling. Automated platforms can process over 2,000 samples per eight-hour shift with minimal technician intervention, representing a 5-10 fold increase in productivity compared to manual column-based methods.
The scalability of silica bead systems extends beyond sample numbers to include flexible sample input volumes, with protocols readily adaptable from 10μL to 1mL of starting material without significant methodology modifications. This flexibility accommodates diverse sample types with varying DNA content, from forensic samples with minimal cellular material to whole blood specimens with abundant nucleic acids. Batch-to-batch consistency in commercial silica bead preparations ensures reproducible performance across different production lots, a critical consideration for regulated environments where validation of extraction efficiency is required for each new reagent lot. The combination of automation compatibility, processing flexibility, and lot consistency makes silica bead technology particularly suitable for laboratories with expanding throughput requirements or fluctuating sample volumes.
Performance Metrics in High-Throughput Applications
DNA yield and purity represent the primary performance indicators for any extraction methodology, with silica bead systems typically achieving 70-90% recovery efficiency across diverse sample types when optimized for specific starting materials. The DNA quality, as measured by A260/A280 ratios, consistently falls within the optimal 1.8-2.0 range, indicating minimal protein contamination, while A260/A230 ratios typically exceed 2.0, reflecting effective removal of chaotropic salts and other chemical contaminants. These purity metrics ensure compatibility with sensitive downstream applications including quantitative PCR and next-generation sequencing, where inhibitors can dramatically impact results. Process consistency, measured through coefficient of variation calculations, generally remains below 10% for yield measurements in automated high-throughput implementations, demonstrating the methodological robustness necessary for large-scale studies.
Extraction speed constitutes another critical performance metric in high-throughput environments, with silica bead protocols typically completing DNA purification in 30-60 minutes for 96-sample batches. This represents a significant improvement over traditional phenol-chloroform extraction requiring several hours and multiple tube transfers. The simplified work flow of bead-based systems, typically involving only four principal steps—lysis, binding, washing, and elution—minimizes processing time while reducing opportunities for sample mix-up or contamination. Throughput efficiency further improves through the elimination of column equilibration, multiple centrifugation, and column transfer steps required in spin-column methodologies, with some automated systems achieving processing times of under 20 minutes for 96-well plates.
Sample Type Considerations and Protocol Optimization
Blood specimens represent one of the most common sample types processed through silica bead systems, with specific protocol modifications recommended for different anticoagulants and storage conditions. EDTA-treated whole blood typically yields higher molecular weight DNA compared to heparinized specimens, as heparin can inhibit downstream enzymatic reactions if not completely removed during extraction. Forensic blood stains often require extended proteinase K digestion and specialized lysis conditions to efficiently release DNA from dried matrices, with yields varying based on stain age and environmental exposure. The forensic blood extraction protocols frequently incorporate additional washing steps to remove hemoglobin and other PCR inhibitors that accumulate in aged specimens.
Formalin-fixed paraffin-embedded tissues present unique challenges for DNA extraction due to protein-DNA crosslinking and nucleic acid fragmentation caused by formalin fixation. Silica bead systems optimized for FFPE samples incorporate extended heating steps at elevated temperatures (often 80-90°C) to reverse formalin-induced crosslinks, combined with vigorous mixing to disperse the paraffin and maximize tissue lysis. The resulting DNA fragments typically range from 100-1000 base pairs, suitable for PCR amplification but challenging for applications requiring high molecular weight DNA. Yield recovery from FFPE blocks varies with fixation time, storage duration, and tissue type, with protocols specifically designed for FFPE animal tissues incorporating additional deparaffinization steps not required for fresh or frozen specimens.
Microbial and environmental samples often contain complex mixtures of organisms with varying cell wall structures, necessitating customized lysis conditions for efficient DNA recovery. Gram-positive bacteria require more rigorous lysis conditions than Gram-negative species, typically incorporating lysozyme treatment and mechanical disruption alongside standard proteinase K digestion. Soil and water samples present additional challenges through the presence of humic acids, heavy metals, and other environmental inhibitors that can co-purify with DNA and interfere with downstream applications. Specialized soil DNA extraction protocols often include polyvinylpyrrolidone or other additives that selectively bind these inhibitors, improving DNA purity while maintaining yield.
Downstream Application Compatibility
Next-generation sequencing applications demand high-quality DNA with minimal contamination from proteins, salts, or organic compounds that can interfere with library preparation or sequencing chemistry. Silica bead-extracted DNA consistently demonstrates excellent performance in NGS applications, with fragment size distribution appropriate for modern sequencing platforms and sufficient purity to ensure efficient adapter ligation and amplification. The absence of significant inhibitors in properly purified samples results in high library conversion rates and uniform sequence coverage, critical metrics for detecting variants in clinical applications or achieving comprehensive representation in metagenomic studies. The reproducible yield and quality from silica bead systems enable accurate normalization of DNA input for library preparation, reducing the need for quantification and dilution steps that introduce variability and increase processing time.
PCR and quantitative PCR applications benefit from the efficient removal of inhibitors that can reduce amplification efficiency or cause complete reaction failure. Hemoglobin from blood samples, humic acids from soil, and urea from forensic specimens represent common inhibitors effectively removed through silica bead purification when protocols include appropriate washing steps. The consistency of silica bead systems ensures uniform PCR performance across sample batches, a critical requirement for diagnostic applications where threshold values must remain stable over time. Real-time PCR amplification efficiency typically exceeds 90% with silica bead-purified DNA, with CT values varying by less than 0.5 cycles between replicate extractions, demonstrating the methodological precision necessary for reliable gene expression analysis or viral load quantification.
Comparison with Alternative Extraction Technologies
Magnetic bead systems share similar binding chemistry with silica beads but employ paramagnetic particles that enable separation using magnetic racks rather than centrifugation or filtration. This distinction provides practical advantages in automation, as magnetic separation integrates more readily with liquid handling platforms than the vacuum manifolds or centrifugation required for traditional silica membranes. However, magnetic systems typically incur higher reagent costs due to the specialized manufacturing processes for paramagnetic silica particles, with per-sample expenses 20-40% greater than conventional silica bead methods. Throughput capacity remains comparable between the technologies, though magnetic systems may demonstrate slight advantages in processing time for very large sample batches due to more rapid separation cycles.
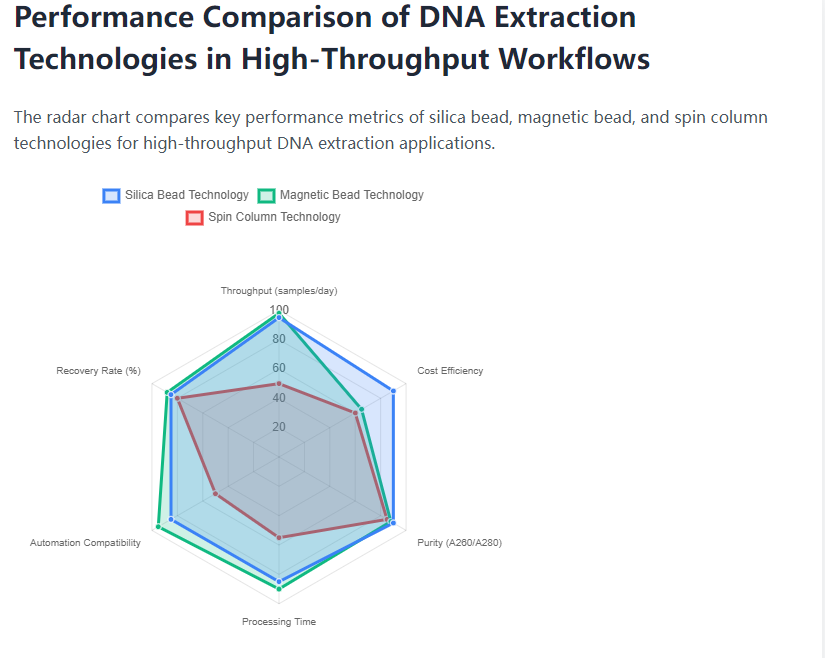
Spin column technology represents the most direct relative to silica bead methods, employing the same binding chemistry but immobilizing silica in membrane form within plastic columns. Column-based systems typically produce DNA with slightly higher molecular weight due to gentler processing conditions, though yields may be lower due to limited binding surface area. The requirement for multiple centrifugation steps makes column technology less amenable to high-throughput automation, though several manufacturers have developed plate-based formats compatible with standard centrifuges. For laboratories processing fewer than 50 samples daily, column systems may provide a cost-effective solution, but the scalability limitations become significant as sample volumes increase beyond this threshold.
Organic extraction methods using phenol-chloroform historically represented the gold standard for DNA purity but have largely been superseded by silica-based methods in high-throughput environments due to safety concerns, processing time, and limited scalability. While organic extraction can produce high-molecular-weight DNA suitable for demanding applications like long-read sequencing, the manual nature of the process, use of hazardous chemicals, and difficulty automating the phase separation steps make this approach impractical for processing more than a few dozen samples simultaneously. The consistent yield and quality of modern silica bead systems, combined with their safety profile and automation compatibility, have established them as the preferred technology for most high-throughput applications.
Implementation Considerations for Laboratory Workflows
Laboratory space and equipment requirements significantly influence the feasibility of implementing silica bead technology, with automated systems typically requiring dedicated bench space for liquid handling platforms and associated equipment. The initial capital investment for automation can be substantial, though the long-term labor savings and increased throughput capacity typically justify this expenditure for laboratories processing more than 10,000 samples annually. For facilities with limited resources, manual processing using multi-channel pipettes and standard laboratory equipment can still achieve respectable throughput of 200-300 samples per technician daily, representing a 3-5 fold improvement over column-based processing. The silica bead extraction kits designed specifically for high-throughput applications typically include pre-mixed reagents and optimized protocols that minimize setup time and reduce opportunities for operator error.
Personnel training requirements vary with implementation scale, with automated systems necessitating specialized technical expertise for operation, maintenance, and troubleshooting. Standard operating procedures should include regular performance verification using control samples to monitor extraction efficiency and identify potential process deviations before they impact experimental results. Documentation systems must track reagent lot numbers, equipment maintenance, and processing parameters to facilitate investigation of any quality issues that may arise. Cross-training technical staff on both automated and manual versions of the protocol ensures continuity of operations during system maintenance or unexpected equipment failure.
Troubleshooting Common Performance Issues
Reduced DNA yield represents one of the most frequent issues encountered with silica bead systems, often resulting from insufficient lysis, incomplete binding, or inefficient elution. Inadequate lysis fails to release DNA from cellular or tissue matrices, particularly challenging samples like plant tissues with robust cell walls or Gram-positive bacteria with complex membrane structures. Optimizing lysis conditions through extended incubation times, increased temperature, or additional enzymatic treatments typically resolves this issue. Incomplete binding frequently stems from incorrect chaotropic salt concentration or pH, while inefficient elution may result from insufficient incubation time or excessive drying of silica beads before elution buffer addition.
PCR inhibition in downstream applications indicates incomplete removal of contaminants during the washing steps of silica bead purification. Common inhibitors include polysaccharides from plant tissues, melanin from hair and skin samples, and hematin from blood specimens, each requiring specific methodological adjustments for effective removal. Additional washing steps with ethanol-containing buffers typically improve purity, though excessive washing may reduce final yield through unnecessary DNA loss. For particularly challenging samples, incorporating inhibitor removal reagents specifically designed to bind problematic compounds often resolves amplification issues without significantly impacting DNA recovery. The implementation of clinical DNA extraction protocols typically includes rigorous validation of inhibition removal to ensure reliable detection of low-abundance pathogens.
Quality Control and Validation Approaches
Implementing comprehensive quality control measures ensures consistent performance of silica bead extraction systems in high-throughput environments, with both process controls and extracted DNA characterization providing valuable performance data. Extraction controls consisting of known quantities of cultured cells or purified DNA processed alongside experimental samples verify that the methodology performs within expected parameters for each batch. Quantification using fluorescence-based methods provides more accurate DNA concentration measurements than spectrophotometric approaches, particularly important for samples with low DNA concentration where accurate normalization is critical for downstream applications. Fragment size analysis through agarose gel electrophoresis or automated electrophoresis systems confirms DNA integrity, a particularly important metric for applications requiring high-molecular-weight DNA.
Method validation establishes performance characteristics including yield, purity, precision, and robustness under standard operating conditions. This process typically involves extracting replicate samples across multiple days by different operators to assess inter-assay and inter-operator variability, with acceptance criteria established based on intended application requirements. For regulated environments, validation must demonstrate that the method consistently produces DNA suitable for its intended use, with documentation supporting claims regarding sensitivity, specificity, and reproducibility. The inclusion of research-grade extraction protocols in validation studies provides benchmark performance data that facilitates comparison across different methodologies and platforms.
Economic Considerations and Return on Investment
The per-sample cost of silica bead extraction varies with scale, reagent source, and degree of automation, typically ranging from $0.50 to $2.00 per sample for high-throughput implementations. This represents a significant reduction compared to column-based systems, which average $2.50 to $5.00 per sample when processing comparable volumes. The economic advantage increases with scale, as silica bead systems benefit more substantially from bulk reagent purchasing and reduced labor requirements. Laboratories should consider both direct reagent costs and indirect expenses including technician time, equipment maintenance, and quality control activities when evaluating total cost per sample, as these factors collectively determine the true economic impact of implementation.
Return on investment calculations for automated silica bead systems typically project payback periods of 12-24 months based on labor savings and increased throughput capacity. The precise economic benefit varies with laboratory-specific factors including sample volume, personnel costs, and current methodology efficiency. Laboratories processing fewer than 5,000 samples annually may find manual silica bead processing more economically viable than automated systems, while facilities exceeding this threshold generally benefit from automation investment. The scalability of silica bead technology enables laboratories to begin with manual processing and transition to automated systems as sample volumes increase, protecting the initial methodology investment while accommodating evolving operational requirements.
Future Directions and Emerging Applications
Microfluidic integration represents the next evolutionary step for silica bead technology, with systems under development that combine extraction, quantification, and amplification in fully automated platforms. These integrated systems promise to reduce processing time to minutes while minimizing sample handling and opportunities for contamination. The miniaturization of extraction chemistry enables significant reagent reduction, potentially decreasing per-sample costs while maintaining or improving yield and purity. Several commercial systems already demonstrate the feasibility of this approach for specific applications, though widespread implementation awaits further technological refinement and validation across diverse sample types.
Application expansion continues as researchers adapt silica bead methodology to increasingly challenging sample types, including ancient DNA specimens, single cells, and circulating tumor DNA from liquid biopsies. Each application presents unique challenges requiring protocol modifications to address specific contaminants, limited starting material, or unusual sample matrices. The fundamental flexibility of the silica bead approach facilitates these adaptations, with binding conditions adjustable through modification of chaotropic salt concentration, pH, and silica surface properties. The ongoing development of food DNA extraction applications demonstrates how silica bead technology continues to evolve to meet emerging needs in quality control and safety testing across diverse industries.
The performance of silica bead DNA extraction kits in high-throughput workflows reflects their optimal balance of efficiency, consistency, and scalability for modern molecular biology applications. The fundamental binding chemistry provides selective nucleic acid capture across diverse sample types, while the bead format enables automation compatibility unmatched by alternative methodologies. Laboratories implementing silica bead systems benefit from reduced processing time, decreased per-sample costs, and improved result consistency compared to traditional extraction approaches. As molecular applications continue to expand in both scale and complexity, silica bead technology remains well-positioned to meet evolving requirements through ongoing refinement and application-specific optimization.
