Forensic DNA analysis represents one of the most demanding applications in molecular biology, requiring exceptional sensitivity, reproducibility, and contamination control. Spin column technology has emerged as the gold standard for forensic DNA extraction due to its balanced performance across critical parameters including yield, purity, and inhibitor removal. This comprehensive examination explores the technical considerations that make spin column methods particularly suited for challenging forensic evidence processing.
The reliability of forensic DNA analysis fundamentally depends on the quality of extracted nucleic acids. According to data from the National Institute of Standards and Technology, approximately 35% of forensic sample processing challenges stem from suboptimal DNA extraction rather than analytical limitations. Spin column systems address these challenges through a combination of chemical principles and physical separation mechanisms that consistently deliver high-quality DNA from diverse sample types.
Fundamental Principles of Spin Column DNA Extraction
Spin column technology operates through a sequence of carefully optimized biochemical reactions and physical separations. The core mechanism relies on the selective binding of DNA to silica-based membranes under specific chaotropic salt conditions, followed by sequential washing to remove contaminants and final elution in low-ionic-strength buffers.
The chemistry underpinning this technology exploits the affinity of DNA molecules for silica surfaces in the presence of high concentrations of chaotropic salts such as guanidine thiocyanate. These salts disrupt hydrogen bonding networks, dehydrate nucleic acids, and enable electrostatic interactions between the DNA phosphate backbone and positively charged silica surfaces.
Cell Lysis and DNA Release Mechanisms
Effective cell lysis represents the critical first step in DNA extraction from forensic samples. Spin column kits typically employ a combination of chemical and enzymatic lysis strategies tailored to specific sample types. Proteinase K digestion at 56°C effectively degrades nucleases and structural proteins while preserving DNA integrity. The lysis buffer composition often includes detergents like SDS that disrupt lipid membranes and chaotropic salts that denature proteins.
The efficiency of DNA recovery heavily depends on complete cell disruption and nuclear lysis. For challenging forensic samples such as bone, teeth, or hair shafts, extended digestion periods up to 24 hours may be necessary to fully release DNA from the mineralized or keratinized matrices. The optimization of lysis conditions directly impacts the success rate of downstream forensic analyses including STR profiling.
Selective DNA Binding to Silica Membranes
The binding phase utilizes the principle that DNA adsorbs to silica surfaces in the presence of high concentrations of chaotropic salts, typically at pH values below the pKa of silanol groups. This creates a positive charge on the silica membrane that attracts the negatively charged DNA backbone. The binding capacity of modern spin columns ranges from 20-50 μg depending on membrane surface area and chemistry.
Binding conditions must be carefully controlled to maximize DNA capture while minimizing co-purification of inhibitors. The optimal binding buffer contains chaotropic salts at concentrations between 4-6 M, maintaining pH around 6.5 to facilitate DNA-silica interactions. Incomplete binding represents a significant source of DNA loss, particularly with low-copy-number forensic samples.
Contaminant Removal Through Sequential Washing
Wash buffers in spin column systems serve to remove proteins, cellular debris, and PCR inhibitors while maintaining DNA binding to the silica membrane. The first wash typically contains chaotropic salts and alcohol to remove contaminants while keeping DNA bound. Subsequent washes with alcohol-based solutions at higher concentrations remove salts and residual contaminants.
The effectiveness of washing steps directly impacts DNA purity, measured by A260/A280 and A260/A230 ratios. Forensic samples often contain complex inhibitors including humic acids from soil, hematin from blood, or indigo dyes from denim that require specialized wash buffers. Incomplete removal of these substances can compromise downstream PCR amplification and STR analysis.
DNA Elution and Recovery Optimization
Elution reverses the DNA-silica binding by disrupting the chaotropic salt environment. Low-ionic-strength buffers such as TE (10 mM Tris-HCl, 1 mM EDTA) or nuclease-free water hydrate the DNA and silica surface, weakening their interaction. The optimal elution pH ranges between 8.0-8.5 to ensure DNA stability while facilitating complete release from the membrane.
Elution efficiency depends on several factors including incubation time, temperature, and buffer volume. Pre-warming elution buffers to 60-70°C can improve DNA recovery by 15-25% for some sample types. The standard elution volume for forensic applications typically ranges from 50-100 μL to concentrate DNA from limited samples while maintaining adequate recovery.
Forensic Sample-Specific Processing Considerations
Forensic evidence presents unique challenges that demand specialized processing protocols within the spin column framework. The diverse nature of forensic samples requires tailored approaches to address variations in DNA content, inhibitor composition, and sample preservation.
Successful DNA extraction from forensic evidence must account for sample degradation, environmental exposure, and minimal biological material. The adaptability of spin column technology to these variables makes it particularly valuable for forensic applications where sample quantity and quality are often limiting factors.
Blood and Bloodstain Evidence Processing
Blood samples represent one of the most common biological materials in forensic casework. The high nuclease activity in blood necessitates rapid processing or proper preservation to prevent DNA degradation. Spin column protocols for blood incorporate specific measures to address heme and iron-containing compounds that can inhibit downstream PCR amplification.
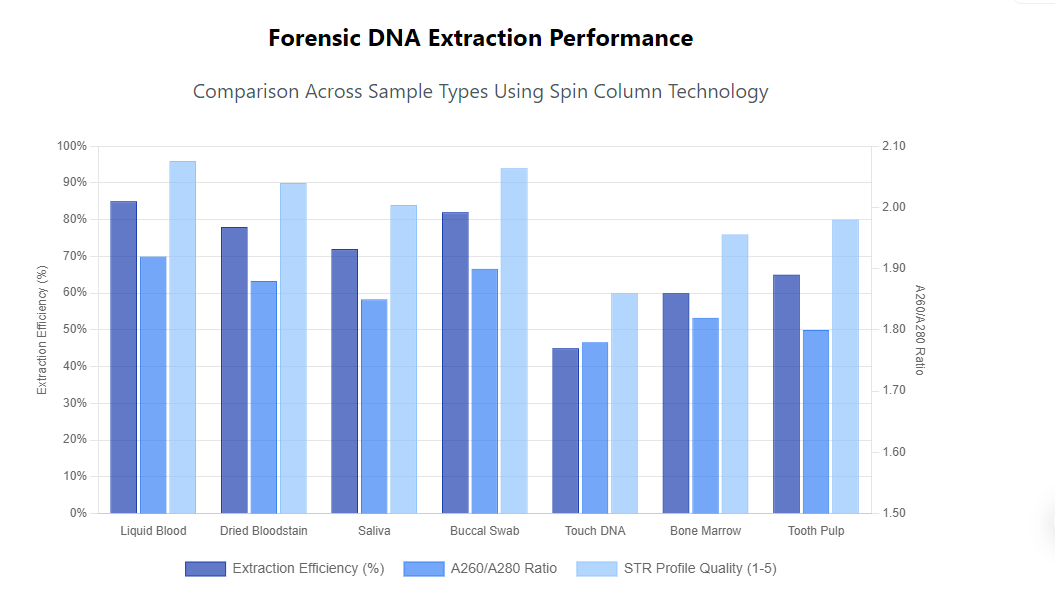
For dried bloodstains, rehydration and extended proteinase K digestion improve DNA recovery from aged samples. The use of specialized forensic DNA extraction kits for blood evidence includes additional washing steps to remove hemoglobin derivatives that can interfere with STR analysis. Recovery rates from bloodstains typically range from 70-90% depending on substrate and storage conditions.
Saliva and Buccal Cell Collection
Saliva evidence presents distinct challenges due to the presence of bacterial DNA and food residues that can complicate analysis. Spin column processing of saliva samples benefits from pre-treatment steps to enrich human DNA, such as selective lysis approaches that target human epithelial cells while minimizing bacterial DNA co-extraction.
Buccal swabs collected as reference samples require optimized protocols to handle the mixture of epithelial cells and bacteria. The addition of carrier RNA or protein during extraction can improve recovery from low-cell-count swabs. Proper swab processing techniques, including thorough rotation during lysis, ensure maximum DNA release from collection devices.
Touch DNA and Trace Evidence Recovery
Touch DNA evidence, containing minimal biological material from skin cells, demands extraction methods with high sensitivity and low contamination risk. Spin column technology adapted for trace evidence often incorporates reduced elution volumes and specialized collection techniques to concentrate the limited DNA available.
The processing of touch DNA requires meticulous attention to contamination prevention throughout the extraction process. Miniaturized spin columns with reduced bed volumes improve recovery from low-template samples by minimizing surface area and subsequent DNA loss. Yield optimization for touch DNA may involve protocol modifications such as extended lysis and carrier addition.
Bone and Tooth Evidence from Decomposed Remains
Skeletal elements represent some of the most challenging forensic samples due to extensive DNA degradation and the presence of PCR inhibitors. Spin column extraction from bone and tooth specimens requires specialized decalcification steps, typically using EDTA, to access the cellular material within the mineralized matrix.
The extended processing time for skeletal remains, often spanning 24-72 hours, necessitates robust spin column membranes that maintain integrity throughout prolonged incubations. DNA recovered from bone typically shows varying degrees of fragmentation, requiring downstream analysis methods adapted to degraded templates. Success rates for bone DNA extraction have improved significantly with optimized spin column protocols, now achieving reliable results even with centuries-old specimens.
Downstream Application Requirements for Forensic DNA
The quality requirements for extracted DNA vary significantly depending on the intended analytical method. Spin column technology must be selected and optimized to meet the specific demands of contemporary forensic DNA analysis techniques.
Modern forensic laboratories employ diverse analytical platforms including capillary electrophoresis for STR profiling, real-time PCR for quantification, and massively parallel sequencing for advanced marker systems. Each application imposes unique requirements on DNA quality, quantity, and purity that influence extraction protocol selection.
Short Tandem Repeat (STR) Analysis Compatibility
STR analysis represents the gold standard in forensic DNA identification, requiring high-molecular-weight DNA with minimal fragmentation. Spin column extraction methods preserve DNA integrity better than many alternative techniques, with average fragment sizes typically exceeding 20 kb when processed from fresh samples. This ensures successful amplification of STR loci ranging from 100-400 bp.
The purity requirements for STR analysis focus primarily on removing PCR inhibitors that can cause allele dropout or imbalanced peak heights. Spin column purification consistently yields DNA with A260/A280 ratios of 1.8-2.0 and A260/A230 ratios above 2.0, indicating minimal protein and organic compound contamination. These purity standards ensure reliable amplification across all STR loci in commercial multiplex kits.
Next-Generation Sequencing Preparation
The adoption of next-generation sequencing in forensic science imposes additional requirements on extracted DNA quality. Sequencing libraries require DNA with minimal single-stranded breaks, chemical modifications, or cross-links that can interfere with adapter ligation and cluster generation. Spin column technology provides DNA suitable for NGS applications when processing conditions are carefully controlled.
DNA fragment size distribution represents a critical parameter for NGS library preparation. Spin column methods can be modified to select for specific size ranges through adjusted binding and washing conditions. The elimination of enzyme inhibitors is particularly important for sequencing applications, as many library preparation steps involve sensitive enzymatic reactions.
Real-Time PCR Quantification Standards
Accurate DNA quantification represents an essential quality control step in forensic analysis. Real-time PCR quantification methods require DNA free of inhibitors that could affect amplification efficiency or interfere with fluorescent detection. Spin column purification effectively removes substances that inhibit DNA polymerases, ensuring reliable quantification results.
The consistency of DNA extraction directly impacts quantification accuracy. Spin column methods demonstrate high reproducibility with coefficients of variation typically below 15% for replicate extractions. This reliability ensures that quantification results accurately reflect the DNA template available for subsequent analysis.
Quality Control and Validation in Forensic DNA Extraction
Forensic applications demand rigorous quality control measures throughout the DNA extraction process. Spin column technology supports comprehensive validation protocols that meet international forensic standards and accreditation requirements.
The implementation of quality systems in forensic DNA extraction ensures result reliability and courtroom admissibility. Spin column methods facilitate the documentation and control of critical parameters that affect DNA quality and analytical outcomes.
Extraction Efficiency and Yield Assessment
Quantifying extraction efficiency requires comparing input biological material to output DNA yield. Spin column methods typically demonstrate efficiency rates of 60-80% for most forensic sample types when optimized for specific applications. Yield assessment includes both total DNA quantification and human-specific measurement to evaluate the effectiveness of human DNA recovery from mixed samples.
The consistency of yield across multiple extractions represents a key validation parameter. Acceptance criteria typically require less than 30% variation in yield for replicate extractions of the same sample type. This reproducibility ensures that casework samples receive consistent processing regardless of when extraction occurs.
Inhibitor Removal and Purity Verification
Verifying inhibitor removal involves testing extracted DNA in downstream applications and monitoring for amplification abnormalities. Spin column methods must demonstrate effective removal of common forensic inhibitors including hematin, humic acids, tannins, and fabric dyes. Purity assessment incorporates spectrophotometric ratios, fluorometric measurements, and functional tests using internal PCR controls.
The establishment of purity thresholds ensures that extracted DNA meets minimum quality standards for forensic analysis. Typical acceptance criteria include A260/A280 ratios between 1.7-2.0 and successful amplification of quantification standards without inhibition indicators. These measures prevent the analysis of compromised DNA that could produce unreliable results.
Cross-Contamination Prevention Measures
Forensic spin column procedures incorporate multiple safeguards against contamination, including physical separations and procedural controls. The use of aerosol-resistant tips, dedicated pre-and post-amplification areas, and reagent blanks monitors potential contamination throughout the extraction process. Spin column technology itself provides a contained system that minimizes opportunities for sample-to-sample carryover.
Validation studies must demonstrate that cross-contamination between samples does not occur at detectable levels. This typically involves processing high-DNA-content samples adjacent to negative controls and verifying the absence of DNA in the controls. The closed nature of spin column systems significantly reduces contamination risk compared to open-tube extraction methods.
Technical Comparison with Alternative Extraction Methods
Spin column technology occupies a specific position in the landscape of DNA extraction methods, offering distinct advantages and limitations compared to alternative approaches. Understanding these differences enables informed selection of extraction methods based on specific forensic application requirements.
The evolution of DNA extraction technologies has produced several competing platforms that challenge the dominance of spin columns in certain applications. Each method demonstrates particular strengths that may better suit specific forensic scenarios or laboratory workflows.
Magnetic Bead-Based Extraction Systems
Magnetic bead technology shares the silica-based binding chemistry with spin columns but employs a different physical separation mechanism. The automation compatibility of magnetic systems offers significant advantages for high-throughput laboratories processing large numbers of similar samples. The closed-tube nature of magnetic extraction reduces contamination risk during processing.
The scalability of magnetic methods makes them suitable for processing both very small and very large sample volumes. However, the higher initial equipment costs and reagent expenses may be prohibitive for smaller laboratories. The performance of magnetic systems for challenging forensic samples has improved significantly, now rivaling spin columns for many applications.
Organic Extraction and Precipitation Methods
Traditional phenol-chloroform extraction continues to offer advantages for certain challenging forensic samples, particularly those with extensive degradation or complex inhibitor profiles. The method's effectiveness stems from the efficient protein denaturation and separation achieved through organic phase partitioning. However, the use of hazardous chemicals and lengthy procedure times limit its application in routine casework.
The DNA yields from organic extraction often exceed those from spin column methods, but purity may be lower due to co-precipitation of inhibitors. The technical skill required and safety concerns have diminished the role of organic extraction in modern forensic laboratories, though it remains valuable for particularly challenging specimens.
Future Directions in Forensic DNA Extraction Technology
The ongoing evolution of DNA extraction technology continues to address the unique demands of forensic applications. Emerging trends focus on increasing sensitivity, reducing processing time, and enhancing automation while maintaining the reliability required for legal proceedings.
Research directions in forensic DNA extraction reflect the changing nature of evidence types and analytical capabilities. These developments build upon the established principles of spin column technology while introducing innovations that address specific limitations.
Microfluidic and Miniaturized Extraction Platforms
Microfluidic devices represent a promising direction for forensic DNA extraction, particularly for minute samples. These systems reduce reagent consumption and processing time while potentially improving recovery through optimized fluid dynamics. The integration of extraction with subsequent analysis steps in lab-on-a-chip formats could revolutionize forensic workflow efficiency.
The development of microfluidic spin column analogs maintains the established silica membrane chemistry while dramatically reducing scale. These systems demonstrate particular promise for processing trace evidence where sample volume represents a significant limitation. The adaptation of these technologies to forensic standards and validation requirements represents an ongoing area of development.
Selective Human DNA Enrichment Techniques
The challenge of mixed samples containing both human and microbial DNA has stimulated development of selective extraction methods. Approaches incorporating human-specific hybridization capture or methylation-based separation show promise for improving the success rate with samples containing minimal human DNA. These methods build upon standard spin column protocols with additional enrichment steps.
The integration of selective technologies with established extraction methods provides a path toward enhanced sensitivity without completely replacing validated protocols. This evolutionary approach maintains the reliability of existing systems while addressing specific limitations encountered with challenging evidence types.
Rapid Processing and Direct Amplification Approaches
The forensic community shows increasing interest in rapid DNA technologies that minimize or eliminate extraction steps. Direct amplification approaches bypass traditional extraction, instead using crude lysates as PCR templates. While these methods offer significant time savings, they remain limited to relatively clean samples with sufficient DNA content.
Spin column technology continues to evolve toward faster processing times while maintaining the purification essential for challenging forensic samples. The development of rapid DNA extraction kits specifically designed for forensic applications reduces processing time to under 30 minutes while maintaining the quality standards required for reliable STR analysis.
